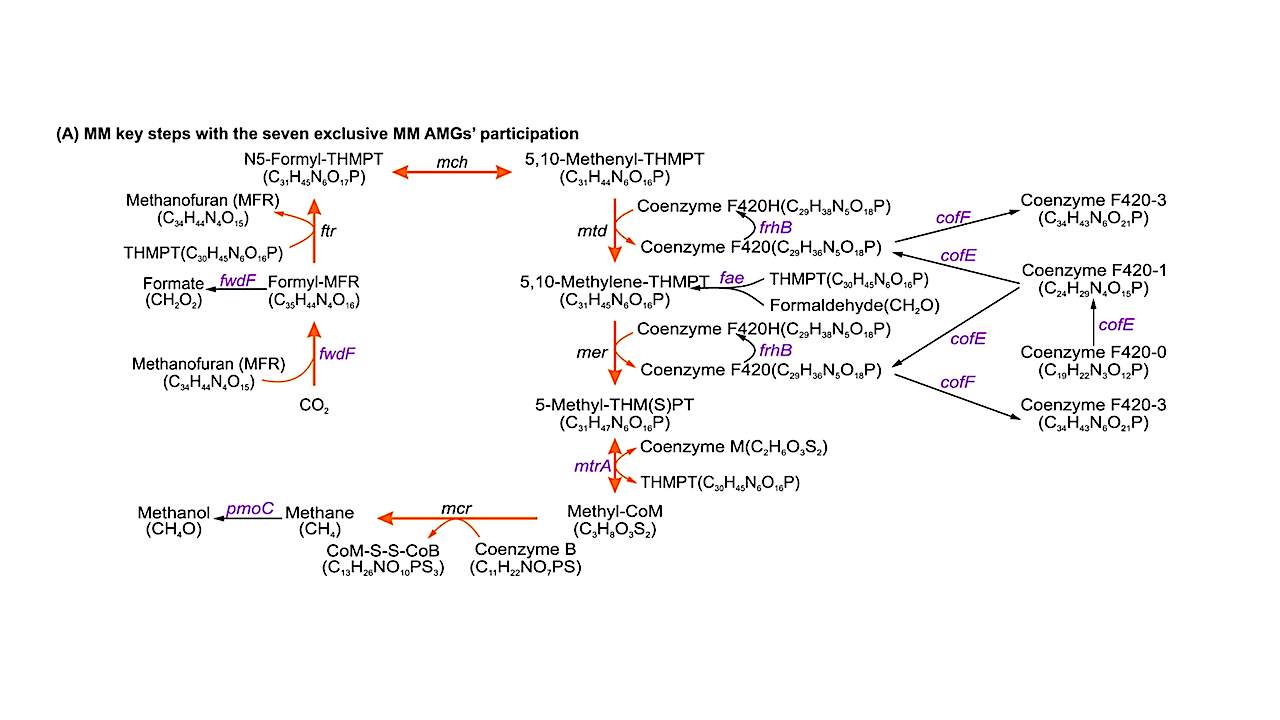
Program for viral participation in key MMP steps by encoding seven AMGs that exclusively participate in MMP. Viruses encoded seven AMGs (fwdF, fae, frhB, cofE, cofF, mtrA and pmoC; in purple text) affecting key steps in both methane production and oxidation. The methanogenesis pathway from CO2 to methane is indicated by orange arrows. Additional information for 17 additional AMGs that may participate in both MMP and other types of metabolic pathways is provided in the Supplementary Figure. S1, S2 and data 5. – Natural correlations
In a new study, scientists have discovered that viruses that infect microbes contribute to climate change by playing a key role in cycling methane, a powerful greenhouse gas, through the environment.
By analyzing nearly 1,000 sets of metagenomic DNA data from 15 different habitats, from various lakes to the inside of a cow's stomach, the microbial viruses have specialized genetic elements that control methane processes, called accessory metabolic genes (AMGs). Depending on where the organisms live, the number of these genes may vary, suggesting that the potential impact of viruses on the environment may vary based on their habitat.
The discovery adds an important piece to better understanding how methane interacts and moves within different ecosystems, said Jiping Zhang, the study's lead author and research associate. Byrd Center for Polar and Climate Research at Ohio State University.
„It's important to understand how microbes drive methane processes,” said Zhang, a microbiologist whose research examines how microbes evolve in different environments. „Microbial contributions to methane metabolic processes have been studied for decades, but research in the viral field is still largely unexplored, and we want to learn more.”
There was study Released today (Feb. 29, 2024) Nature Communications.
Viruses have helped fuel all of Earth's ecological, biochemical, and evolutionary processes, but it's only relatively recently that scientists have begun to explore their relationships with climate change. For example, methane is the second largest driver of greenhouse gas emissions after carbon dioxide, but is mostly produced by single-celled organisms called archaea.
„Viruses are the most abundant biological entity on Earth,” said Matthew Sullivan, a professor of microbiology and co-author of the study at the Center for Microbial Sciences at Ohio State. „Here, we expanded what we know about the effects of methane cycling genes by adding them to the long list of virus-encoded metabolic genes. Our team sought to answer how much 'microbial metabolism' viruses actually manipulate during infection.
Although the important role microbes play in accelerating atmospheric warming is now well known, little is known about how methane metabolism-related genes encoded by viruses that infect these microbes affect their methane production, Zhang said. Solving this mystery led Zhang and his colleagues to spend nearly a decade collecting and analyzing microbial and viral DNA samples from unique microbial reservoirs.
Lake Virana, part of a protected nature reserve in Croatia, was one of the most important locations the team chose to study. Inside the methane-rich lake sediment, the researchers found an abundance of microbial genes that affect methane production and oxidation. In addition, they discovered different viral communities and discovered 13 types of AMGs that help regulate their host's metabolism. Although there is no evidence that these viruses directly encode methane metabolism genes, the potential impact of viruses on methane cycling may vary depending on their habitat, Zhang said.
Overall, the study revealed that a large number of methane-metabolizing AMGs were found to be more abundant within host-associated environments such as the cow stomach. With cows and other livestock responsible for 40% of global methane emissions, their work suggests that the complex relationship between viruses, organisms and the environment may be more intricately connected than scientists once thought.
„These findings suggest that the global impacts of viruses are underestimated and should be given more attention,” Zhang said.
Although it is unclear whether human activities have influenced the evolution of these viruses, the team hopes that new insights gleaned from this work will raise awareness of the power of infectious agents that inhabit all life on Earth. However, more experiments are needed to learn more about the internal mechanisms of these viruses and their contributions to the Earth's methane cycle, Zhang said, especially as scientists work on ways to mitigate microbial-driven methane emissions.
„This work is an initial step toward understanding the virulence impacts of climate change,” he said. „We still have a lot to learn.
This work was supported by the National Science Foundation, the Croatian Science Foundation, the Gordon and Betty Moore Foundation, the Heising-Simons Foundation, the European Union, and the US Department of Energy. Co-authors include Jingjie Du of Ohio State, Stefan Kostlebacher and Petra Bjevac of the University of Vienna, and Sandy Orlick of the Ruger Boskovic Institute.
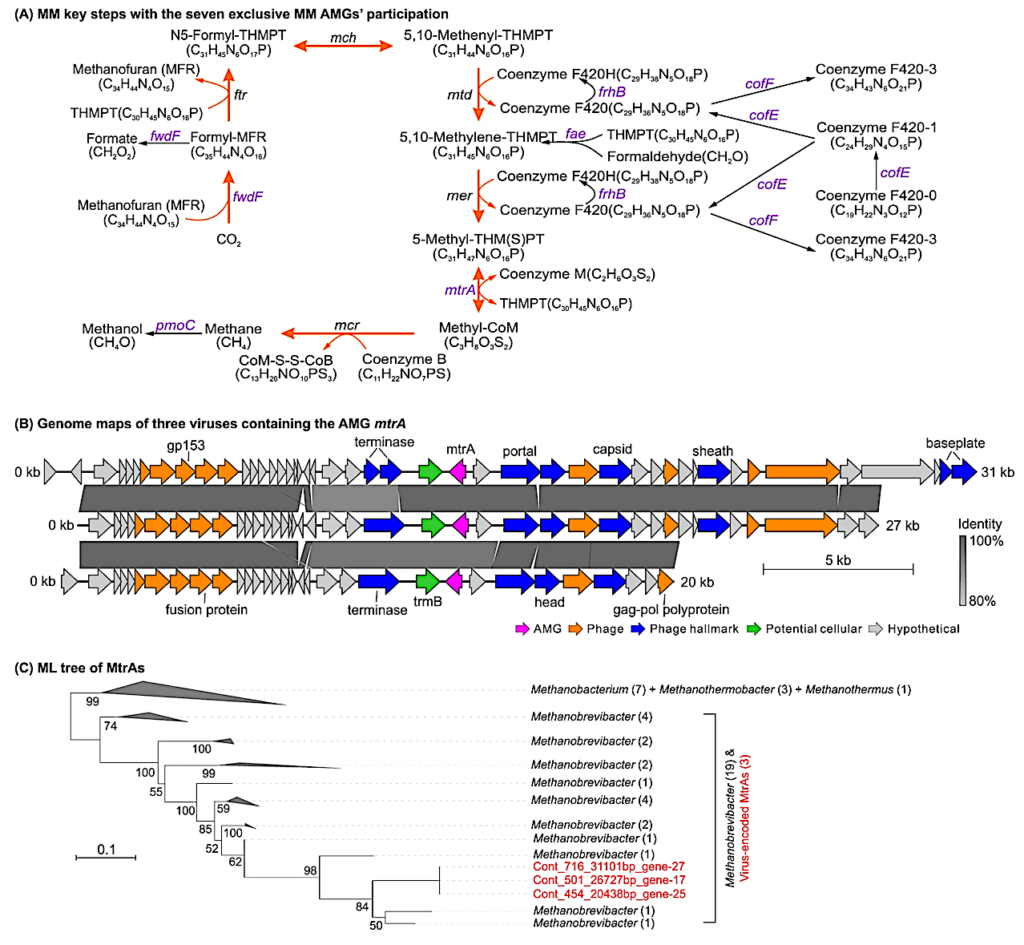
Program for viral participation in key MMP steps by encoding seven AMGs that exclusively participate in MMP. Viruses encoded seven AMGs (fwdF, fae, frhB, cofE, cofF, mtrA and pmoC; in purple text) affecting key steps in both methane production and oxidation. The methanogenesis pathway from CO2 to methane is indicated by orange arrows. Additional information for 17 additional AMGs that may participate in both MMP and other types of metabolic pathways is provided in the Supplementary Figure. S1, S2 and Data 5. B Genome maps of three viral contigs carrying the AMG mtrA gene. All three viral contigs belonged to the same viral population (with 97.4–97.8% genetic identity to each other) and carried the same mtrA gene. CheckV was used to estimate host-virus boundaries and remove potential host fragments in the virus contig. Genes were marked with five colors to illustrate AMGs (purple), phage genes (orange), phage hallmark genes (blue), potential cellular genes (green) and putative protein genes (grey). C Phylogenetic tree of viral and microbial mtrA genes. The tree was inferred using the maximum likelihood method with protein sequences. Parametric bootstrap values (expressed as percentages of 1,000 replicates) are shown at branch points. The scale indicates the distance of 0.1 substitutions per position in the alignment. Viral and microbial MtrA sequences are indicated in red and black, respectively. Numbers in parentheses indicate the number of protein sequences assigned to each group. The full phylogenetic tree (without collapsed groups) is presented in Supplementary Figure S5A. Gene maps and phylogenetic trees for the other six unique MM AMGs ( pmoC , fwdF , fae , cofE , cofF , and frhB ) are presented in Supplementary Fig. S3 and S5B-G. MM, methane metabolism; MMP, methane metabolic pathway.
Viral ability to modulate microbial methane metabolism varies with habitatNature Communications
Astronomy, climate change,

„Oddany rozwiązywacz problemów. Przyjazny hipsterom praktykant bekonu. Miłośnik kawy. Nieuleczalny introwertyk. Student.
