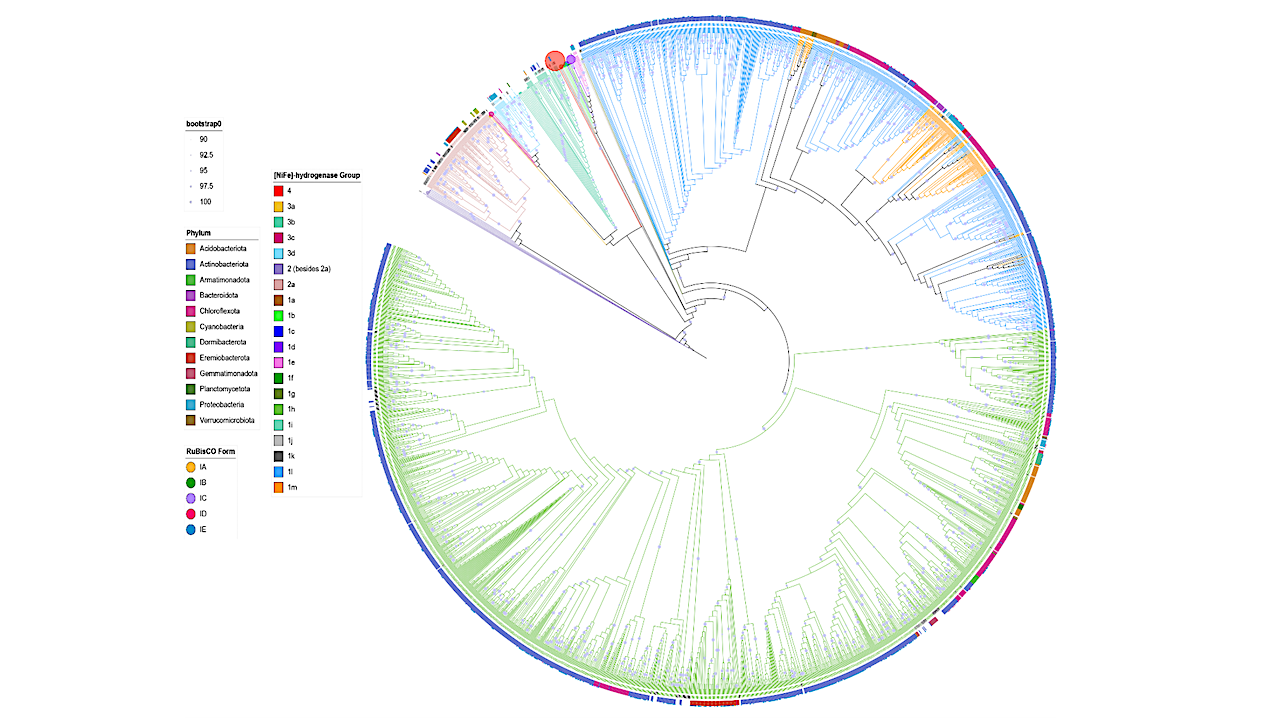
Maximum likelihood phylogenetic tree [NiFe]-HydB and hydrogenase gene sequences obtained from our MAGs (n = 2433), along with reference sequences obtained from previous phylogenetic analysis. Branch and reference gene labels are colored according to group [NiFe]- Hydrogenase. Bootstrap values are depicted as filled circles on branches > 90%, the size reflecting the value and 1000 ultrafast bootstrap iterations were used. The phyla of MAGs collected in this study is shown in the color-coded outer ring. In cases where RuBisCO large subunit gene/s co-occur within these genes, the proportion of motifs present is indicated by outer pie charts. — biorxiv.org
biorxiv.org
Background: Endolithic sites provide a final refuge, providing buffering conditions for microbes inhabiting rock spaces. Nevertheless, the survival and growth strategies of Antarctic endolithic microbes living in Earth’s driest and coldest deserts are virtually unknown.
Results: From 109 endolithic microbes, 4,539 metagenome-assembled genomes were generated, of which 49.3% were novel candidate bacterial species. We present evidence that trace gas oxidation and atmospheric chemistry may be widespread strategies that support metabolic function and the persistence of these ecosystems at the edge of life and habitable limits.
Conclusions: These results represent the foundation for unraveling adaptability at the edge of Earth’s stability and in dry Earth-like planetary bodies such as Mars.
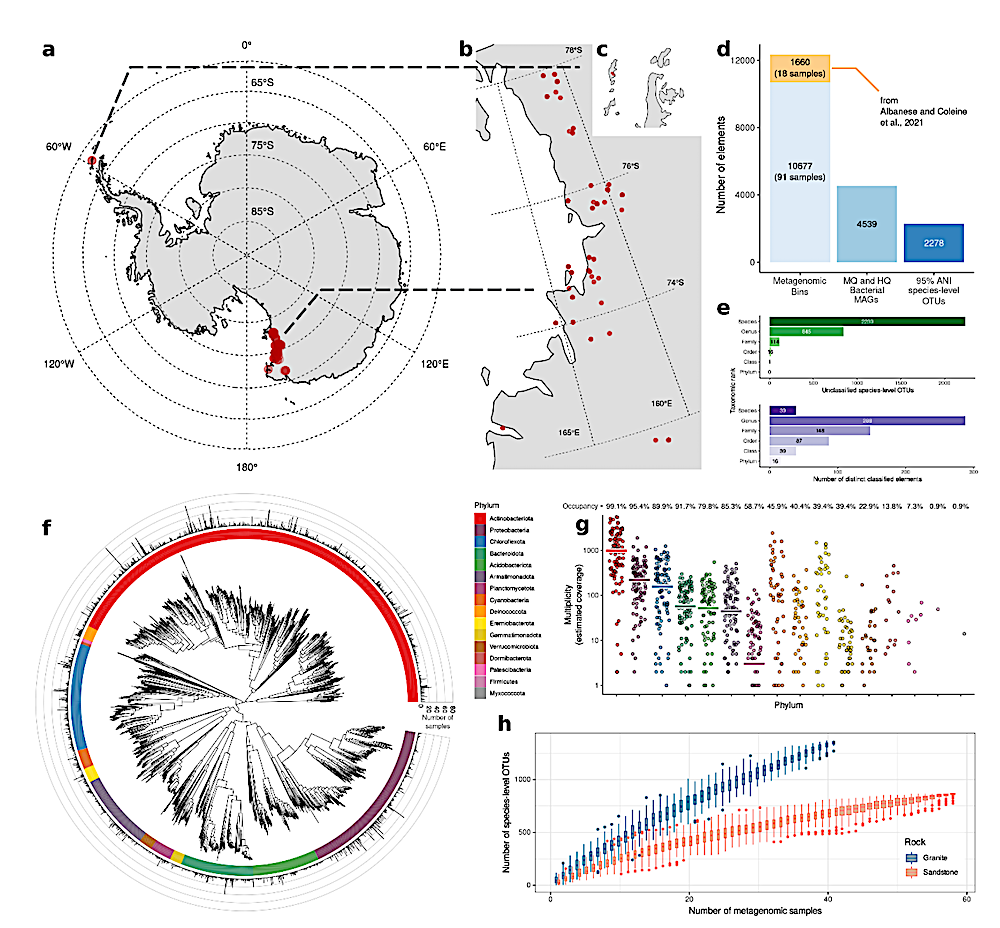
ac, Map of Antarctica (a) and sample sites (Victoria Land, b; Peninsula, c) (red dots). d, Number of MAGs and their qualitative classification. e, Upper bar plot: number of unclassified OTUs. Bottom bar plot: number of species, genera, families, orders, classes and phyla. f, Phylogenetic tree of 2,278 OTUs constructed from multiple sequence alignment of 120 GTDB marker genes. The barplot in the outer circle represents the number of samples in which each OTU was detected. g, Phylum-level mesh screen multiplex for each sample, indicating sequence coverage. Horizontal lines indicate mean values. Occupancy value indicates the percentage of samples containing the underlying phylum. h, Number of OTUs as a function of number of rock samples. — biorxiv.org
doi: https://doi.org/10.1101/2023.07.30.551190
https://www.biorxiv.org/content/10.1101/2023.07.30.551190v1biorxiv.org
Astronomy

„Oddany rozwiązywacz problemów. Przyjazny hipsterom praktykant bekonu. Miłośnik kawy. Nieuleczalny introwertyk. Student.
